Acceleration of First-Principles Statistical Thermodynamics Framework abICS Using an On-Lattice Neural Network Model and Active Learning
PI of Joint-use project: S. Kasamatsu
Host lab: Supercomputer Center
Host lab: Supercomputer Center
In 2019, ab Initio Configuration Sampling toolkit, or abICS for short, was developed as an ISSP Software Advancement Project for directly combining first-principles relaxation and total energy calculations with replica exchange Monte Carlo (RXMC) sampling [1]. The motivation was that effective models such as cluster expansion, which are used to speed up the usual Metropolis Monte Carlo sampling for analyzing order/disorder in alloy systems, are limited in describing complex many-component systems with sufficient accuracy. Our aim at that time was to enable direct sampling on first-principles energies by employing highly parallel sampling methods such as RXMC in combination with massively parallel supercomputing resources. A few years later, some of the authors found that an on-lattice neural network model overcomes many issues found in previous effective models if sufficient training data is provided in an active learning setting [2,3]. This led to the proposal of last year’s Software Advancement Project, which we outline in this report.
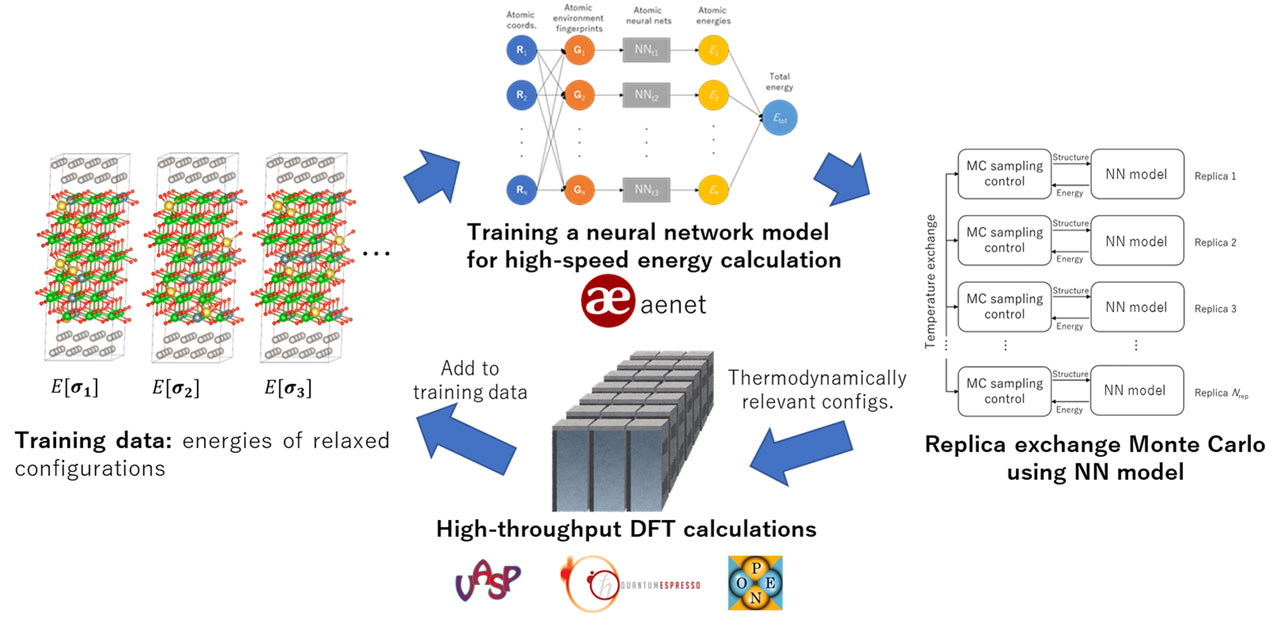
The abICS framework developed in this project uses Python as a glue language to piece together the necessary components (Fig. 1). It is registered on PyPI (Python Package Index; pypi.org) and can be installed easily in most environments through the command “pip install --user abics”. A typical use case is outlined as follows:
- Random configurations on a lattice are relaxed and their total energies are calculated using the first-principles code of choice. We support VASP, Quantum Espresso, and OpenMX.
- A neural network model is trained on data from step 1 to predict relaxed energies from configurations on the non-relaxed lattice. We currently use aenet code [4] for the neural network training.
- RXMC or population annealing MC (PAMC) calculation is performed using the neural network model from step 2. Two sampling modes are available: canonical sampling with a fixed number of atoms or grand canonical sampling which allows for changes in the composition. For the neural network evaluation, we provide a file IO-based interface to aenet, and we also provide an interface to aenet-lammps [5] python interface which does not rely on file IO and is thus usually faster.
- A subset of samples from step 3 are relaxed and their total energies are calculated using first-principles calculation. If the results deviate considerably from the neural network prediction, the data is added to the training data set and the procedure is repeated from step 2.
The overall procedure is controlled by an input file in TOML format, which is an easy-to-read software configuration format that is being used in many projects [6]. The parameters for the first-principles calculations and neural network training/evaluation are controlled by separate files following formats of the specified solver.
We believe that abICS will be a game-changer in modeling order/disorder in many-component crystalline systems as we have already demonstrated for partially hydrated Sc-doped BaZrO3 [3]. Please do not refrain from contacting us if you find any difficulties in using or extending this software.
References
- [1] S. Kasamatsu and O. Sugino, J. Phys.: Condens. Matter 31, 085901 (2019).
- [2] S. Kasamatsu et al., J. Chem. Phys. 157, 104114 (2022).
- [3] K. Hoshino, S. Kasamatsu et al., Chem. Mater. 35, 2289 (2023).
- [4] N. Artrith et al., Phys. Rev. B 96, 014112 (2017).
- [5] M. S. Chen et al., J. Chem. Phys. 155, 074801 (2021).
- [6] https://github.com/toml-lang/toml