Functional Characterization and Study on the Molecular Mechanism of Ion-transporting Rhodopsins
Inoue Group
Rhodopsin is photo-receptive heptahelical transmembrane protein in which a retinal chromophore is covalently bound to a conserved lysine residue in the seventh transmembrane helix (TM7), and animal and microbial rhodopsin families are known so far. Animal rhodopsins are present in animal retina to transfer visual signals to brain and are also related to non-visual light sensing in various tissues. Microbial rhodopsins show diverse functions: light-driven ion pump, light-gated ion channel, phototactic sensor, light-dependent regulation of gene expression and enzyme, and so on. Both types of rhodopsins are being widely used in optogenetics to control various cellular events, such as neural activity, gene expression, dynamic protein localization, and so on, by light.
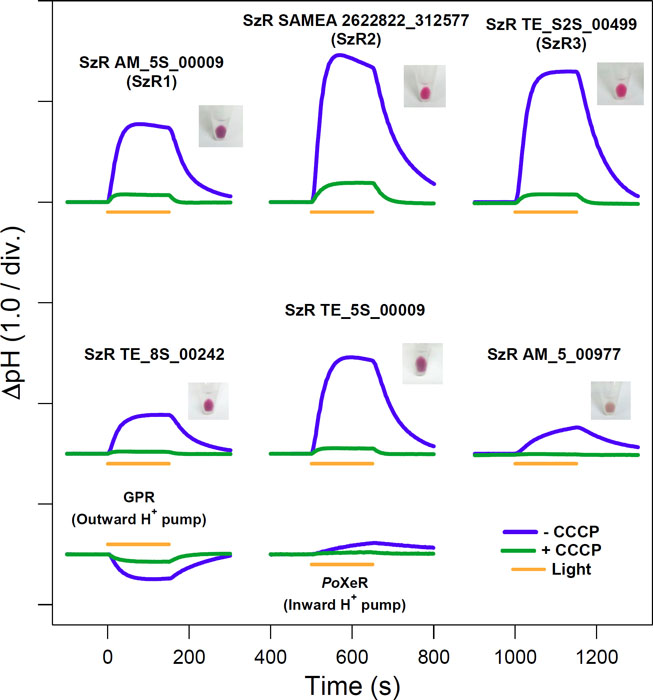
In 2020, we reported the functional characterization of schizorhodopsin (SzR) which was found in metagenome assembled genomes of Asgard archaea which is thought to be the closest living prokaryote to the last common ancestor of eukaryotes [1]. The ion-transport activity assays revealed that SzRs function as light-driven inward proton (H+) pumps [2]. The inward H+ transport of SzRs are significantly higher than that of another inward H+ pumping rhodopsin, xenorhodopsins (Fig. 1). The high-speed atomic force microscopy revealed the trimeric structure of SzR in lipid bilayer. The photocycle of SzR in which K, L, L/M and M intermediates are included was determined by the laser flash photolysis spectroscopy. The transient absorption change of a pH indicator (cresol red) indicated that the substrate H+ is released to the cytoplasmic milieu and taken up from the extracellular side on the M-accumulation and decay, respectively. This suggests that no metastable trapping of H+ released from retinal Schiff base (RSB) occurs on any amino acid residues, and it was supported by the low-temperature Fourier transform infrared spectroscopy. This is quite different from XeR, in which protonation of the proton accepting aspartate on the cytoplasmic side is involved in H+ transfer from the RSB to the cytoplasmic milieu. A comprehensive amino-acid mutation experiment suggested that Cys75 and Glu81 in the transmembrane helix (TM) 3 and Asp184 in TM7 is critical for the inward H+ transport of SzR.
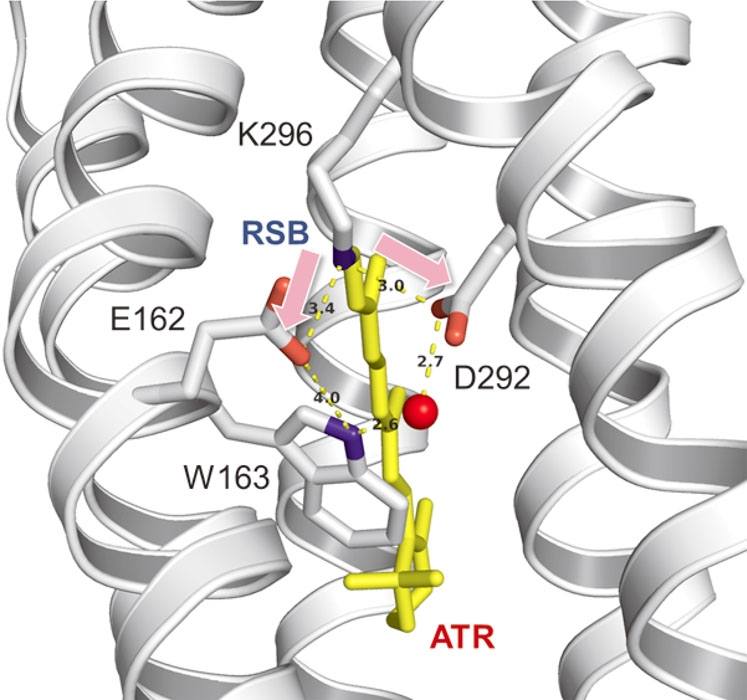
Channelrhodopsins (ChRs) are light-gated cation channels which nonspecifically transports various cations according to the electrochemical potentials of cells. In order to reveal the cation transport function of ChR, we investigated a mutant of a member of ChR, C1C2, in which a tryptophane residue in TM3 was substituted with phenylalanine (C1C2 W163F). Trp163 of C1C2 is highly conserved in most microbial rhodopsins, but its relationship to the channel function is not known. C1C2 W163F showed greatly attenuated channel activity, while outward proton pumping activity became more prominent. This result indicates that Trp163 plays a critical role in suppressing the outward proton pumping function and increasing channel activity by regulating the branching ratio of proton transfer to the two counterions Glu162 and Asp292 respectively (Fig. 2) [3].
We also reported the theoretical and experimental studies on the Cotton effect observed in the visible CD spectrum of sodium pump rhodopsin (KR2) [4]. KR2 shows a visible CD spectrum whose peak is significantly red-shifted from that of its absorption spectrum. The transition density fragment interaction (TDFI) method quantitatively revealed that this shift is caused by the excitonic coupling between retinal chromophores in pentamer of KR2 [4].
References
- [1] P.-A. Bulzu, A.-Ş. Andrei, M. M. Salcher, M. Mehrshad, K. Inoue, H. Kandori, O. Béjà, R. Ghai, and H. L. Banciu. Nat. Microbiol. 4, 1129 (2019)
- [2] K. Inoue, S. P. Tsunoda, M. Singh, S. Tomida, S. Hososhima, M. Konno, R. Nakamura, H. Watanabe, P.-A. Bulzu, H. L. Banciu, A.-Ş. Andrei, T. Uchihashi, R. Ghai, O. Béjà, and H. Kandori, Sci. Adv. 6, eaaz2441 (2020)
- [3] Y. Nagasaka, S. Hososhima, N. Kubo, T. Nagata, H. Kandori, K. Inoue, and H. Yawo. Biophys. Physicobiol. 17, 59 (2020)
- [4] K. J. Fujimoto and K. Inoue, J. Chem. Phys. 153, 045101 (2020).