The Structural Study on MicrobialRhodopsins and the Characterization of New Microbial Rhodopsin Families
Inoue Group
Rhodopsin is photo-receptive heptahelical transmembrane protein in which a retinal chromophore is covalently bound to a conserved lysine residue in the seventh transmembrane helix (TM7), and animal and microbial rhodopsin families are known so far. Animal rhodopsins are present in animal retina to transfer visual signal to brain and are also related to non-visual light sensing in various tissues. Microbial rhodopsins show diverse functions: light-driven ion pump, light-gated ion channel, phototactic sensor, light-dependent regulation of gene expression and enzyme, and so on. Both types of rhodopsins are being widely used in optogenetics to control various cellular events, such as neural activity, gene expression, dynamic protein localization, and so on, by light.
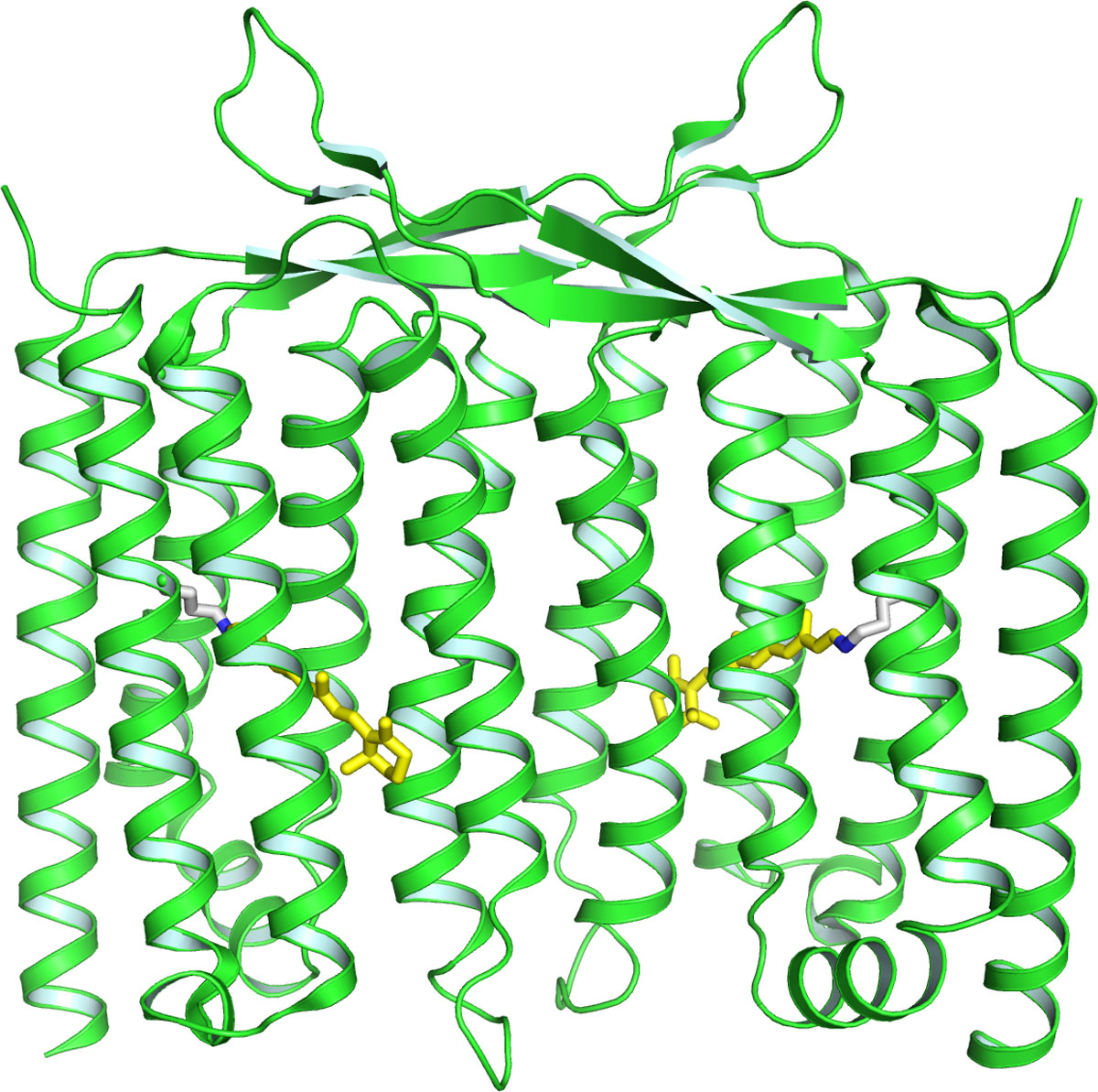
Recently, we discovered the third class of rhodopsin, heliorhodopsin (HeR), which is evolutionally distinct from both of microbial and animal rhodopsins. In 2019, we reported the three-dimensional structure of HeR at atomic level by the x-ray crystallographic analysis (Fig. 1) [1]. It revealed that HeR forms dimers and a fenestration is present near the β-ionone ring of the retinal chromophore. The biochemical experiment suggested that this fenestration is used for the uptake of the retinal chromophore from the environment. This molecular character unique for HeR is considered to be physiologically important for microorganisms having HeR without retinal synthetic genes. Also, the bioinformatic analysis suggested that, while HeR is widely distributed in many gram-positive bacteria, no gram-negative bacteria do not have HeR in their genomes. Because many typical microbial rhodopsins are used by many gram-negative bacteria, the absence of HeR in them is a highly unexpected fact suggesting that the physiological function of HeR would be related to the structure of the cellular membrane of gram-positive bacteria without outer membranes. The study on the ultrafast photoisomerization process of the retinal chromophore in HeR revealed that its isomerization process is similar to those of typical microbial rhodopsins despite of considerably different amino acid residues surrounding the chromophore. A convergent evolution may have occurred in typical microbial rhodopsins and HeR to achieve efficient retinal isomerization with high quantum yields.
The three-dimensional atomic structure of Gloeobacter rhodopsin (GR) was solved by the x-ray crystallographic analysis. The electron microscopic observation revealed that GR exists as a pentamer, and the structural detail obtained in our study will provide new insights to understand the mechanism of oligomerization of membrane protein.
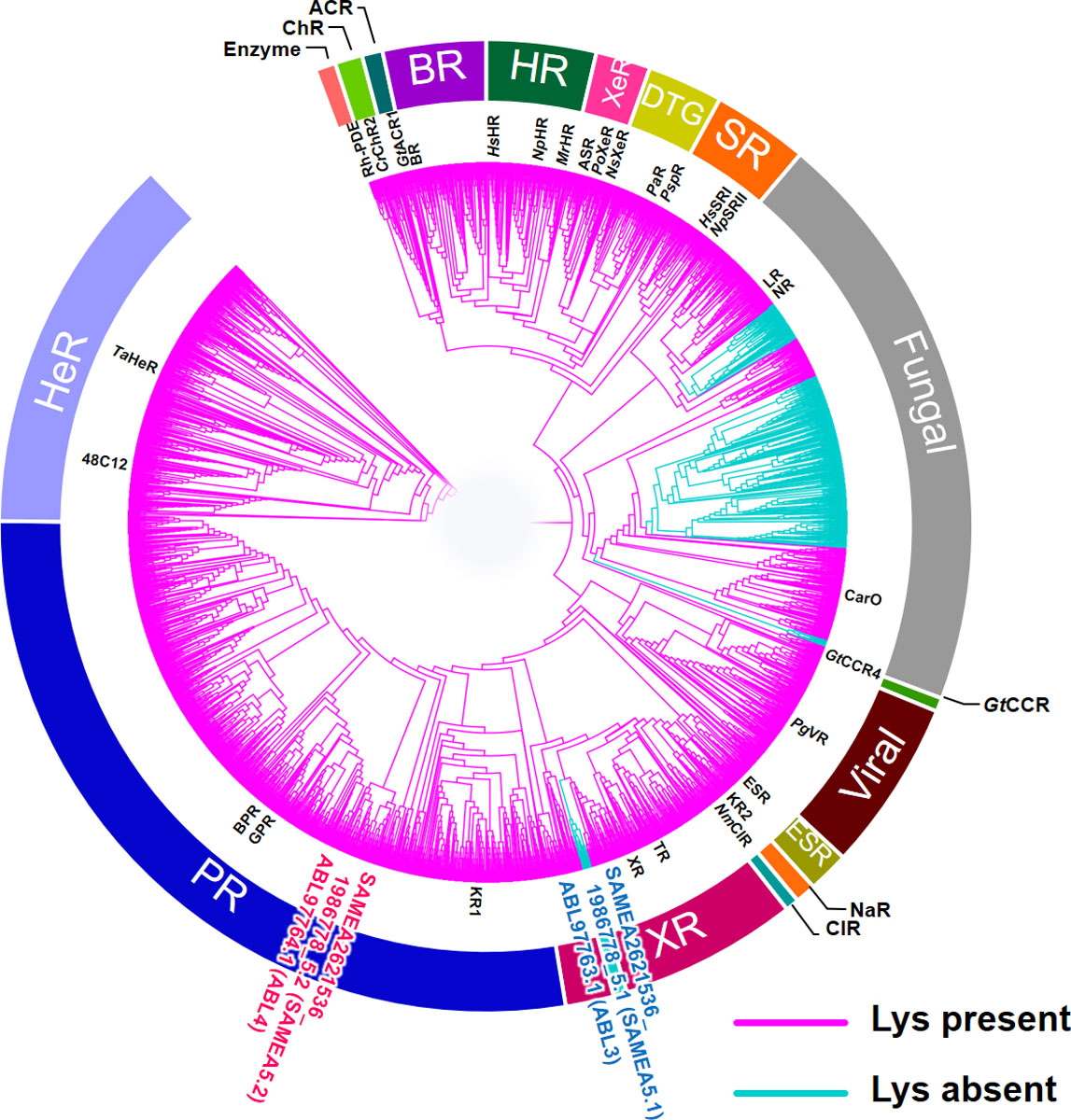
A new type of rhodopsin showing unique isomerization process and microbial rhodopsin without retinal binding lysine (Rh-noK) were also identified from bacteria [2]. The former has a new type of amino acid residues in the TM3 consisting of threonine, alanine and threonine called the “TAT motif” at the position where typical proton pumping rhodopsin has the DTD (aspartic acid-threonine-aspartic acid) or DTE (aspartic acid-threonine-glutamic acid) motifs. In the study of Rh-noK, we revealed that ca. 10% of 5,558 microbial rhodopsins do not have the lysine residue in TM7 which typically is used to bind the retinal chromophore by making a covalent Schiff base linkage (Fig. 2). A colored protein was formed by introducing a lysine residue at this position, and, interestingly, additional introduction of an aspartate functioning as a counterion to the protonated retinal Schiff-base recovered the light-driven outward proton pumping function. This suggests that the Rh-noK protein was recently evolved from natural outward proton pumping rhodopsin, and most of structural elements important for the pump function has been retained even after the lack of the retinal binding lysine and counterion aspartate. The physiological roles of TAT rhodopsin and Rh-noK have not been revealed yet, and they will be investigated in the near future.
References
- [1] W. Shihoya, K. Inoue et al., Nature 574, 132 (2019).
- [2] C. Kataoka, K. Inoue et al., J. Phys. Chem. Lett. 10, 5117 (2019).